Data Insights:
- Users need to provide a comma-separated file in csv format containing lncRNA-miRNA sequence and class information shown as:
- Exploratory sequence analysis engine illustrates characteristics of dataset such as minimum/maximum/average sequence length and sequence-to-label distribution.
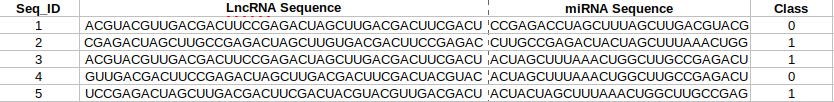
Training Mode
- Users need to provide a csv file containing lncRNA-miRNA sequence pairs and class information.
- They have the freedom to choose the degree and stride values for the generation of desired higher order residues (kmer), distribution of residues by mentioning number of residues to consider from start-end region of lncRNA-miRNA-sequence pairs for optimal sub-sequence generation, number of folds, number of epochs and learning rate to train the model from scratch.
- At the end of training, users can download performance related artifacts to analyze the model behavior.
Prediction Mode:
- Users can upload a csv file of test sequences and perform inference using pre-trained benchmark model.
- At the end of inference, csv artifact containing model predictions can be downloaded to analyze the performance of model.